AI, -OMICs and breakthroughs in organoid vascularization - OD #11
Check out the latest use cases for organoids 🧬
Highlights of this edition:
Microfluidic Organoids + AI Optimize Drug Scheduling for Combination Therapy
Organoid Library + Multi-Omics Uncover Translatable Targets in Pancreatic Cancer
Our GPT Organoids to help you find the best 3D model for your research
More details below 👇
Welcome to Organoids Digest, your monthly update on organoids, organ-on-a-chip, and new approaches methologies (NAMs).
We wish you an enjoyable read ❤️
Article: Co-development of mesoderm and endoderm enables organotypic vascularization in lung and gut organoids
Miao, Pek, Tan et al | August 7, 2025 | Cell
A new protocol for co-developing mesoderm and endoderm within a single spheroid enabled the creation of vascularized lung and intestinal organoids, advancing the field of organoid vascularization.
Context:
In organoid research, vascularization is a hot and rapidly advancing topic. If you have been following recent news on this matter, you may have stumbled upon Abilez et al. (Stanford) study showing that human pluripotent stem cells can be directed to form cardiac and liver organoids with intrinsic vasculature in a single differentiation protocol, without adding endothelial cells later. Building on this momentum, Miao, Pek, Tan, and colleagues (University of Cincinnati College of Medicine) now report vascularized gut and lung organoids, underscoring how quickly the field is expanding across multiple organ systems.
Here’s what the researchers did:
Used human iPSCs to generate spheroids where mesoderm and endoderm co-differentiate, rather than sequential lineage induction.
Tuned BMP signaling to balance mesoderm/endoderm proportions, producing both epithelial progenitors and endothelial/mesenchymal lineages.
Added angiogenic cues (VEGFA, ANG1) during organogenesis to support vessel formation.
Characterized cell types with scRNA-seq, imaging, spatial transcriptomics, and functional assays (barrier integrity, endothelial identity).
Transplanted organoids under the kidney capsule of mice to test in vivo vascular integration and maturation.
Modeled FOXF1 congenital disorders, where abnormal endothelial–epithelial crosstalk disrupts lung and gut development.
Key findings:
Generated vascularized lung and intestinal organoids (vHLPOs, vHIOs) with organ-specific endothelium and mesenchyme resembling human fetal tissues.
Demonstrated functional barrier properties of lung vs intestinal ECs, matching their organ roles (tight vs leaky barriers).
Showed in vivo perfusion and host circulation integration upon transplantation, enhancing maturation and epithelial diversity.
Identified organ-specific ligand–receptor cues (e.g., WNT, semaphorins) that direct endothelial specialization.
Successfully modeled FOXF1-associated congenital vascular disorders, capturing both vascular and epithelial defects.
Why it matters:
This study shows that multilineage differentiation (mesoderm + endoderm) can lead to physiologically relevant organotypic vasculature in organoids. Organoids that self-develop vasculature not only improve fidelity for development and disease modeling, but also open paths to regenerative medicine and personalized therapy for vascular-related congenital and acquired diseases.
Article: Machine learning-assisted exploration of multidrug-drug administration regimens for organoid arrays
Yakavets et al | July 30, 2025 | Science Advances
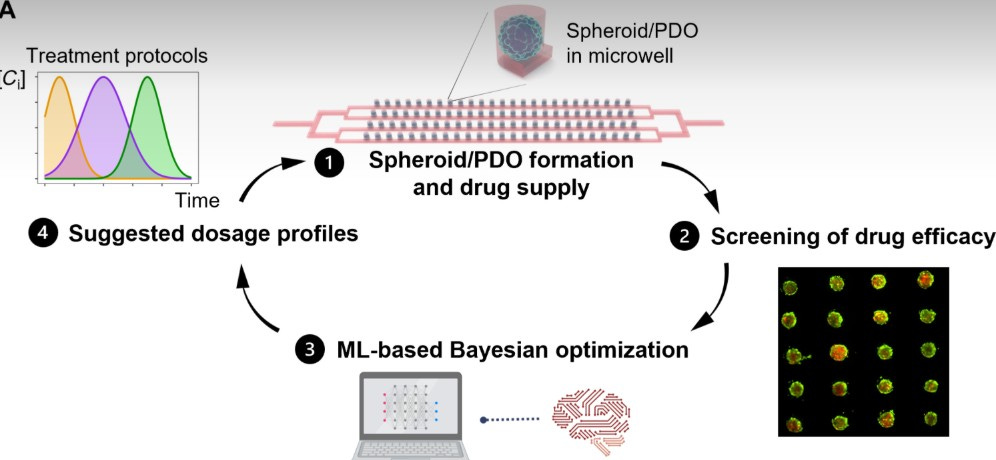
A closed-loop microfluidic organoid platform, integrated with Bayesian optimization, demonstrated how dosing schedule (concurrent vs sequential) can fundamentally change combination therapy outcomes, revealing when drug exposure can be cut dramatically without losing efficacy.
Context:
Determining the optimal doses × order × timing for multidrug regimens is highly time-consuming with classical methods, which typically rely on empirical fixed-dose simultaneous combinations. By merging organoid-based tumor models with machine learning, the study sought to discover clinically meaningful regimens faster and with fewer samples.
Here’s what the researchers did:
Built a microfluidic array chip (1,200 microwells) seeded with uniform 3D breast cancer spheroids and patient-derived organoids (PDOs) in an ECM-mimetic hydrogel (EKGel) under physiologic flow.
Applied a closed-loop Bayesian optimization algorithm (Gryffin) to search drug dose, sequence, and duration space efficiently, prioritizing Bliss synergy first and viability second.
Tested two models: MCF-7 spheroids with the clinical triple (5-FU + DOX + CPA) and breast PDOs with targeted agents (olaparib + IBET-762).
Quantified effects via LIVE/DEAD viability, proliferation (Ki-67), and DNA damage (γH2AX).
Key findings:
In MCF-7 spheroids, an optimized sequential schedule achieved comparable efficacy to concurrent dosing but with a ~75% reduction in total drug exposure.
In breast cancer PDOs, concurrent olaparib + IBET-762 achieved strong synergy (CI ≈ −0.21, viability ≈ 42%), whereas all sequential orders failed, showing that synergy was timing-dependent.
The optimization loop identified strong regimens after only ~10% of the search space was tested, proving high sample efficiency.
Why it matters:
This work shows how organoid-based microfluidics + AI can untangle the complexity of drug scheduling, demonstrating that some regimens allow dose-sparing sequential strategies, while others demand strict concurrency to preserve synergy. It illustrates a path toward patient-specific, empirically guided scheduling using new approaches methodologies.
Article: A pancreatic cancer organoid biobank links multi-omics signatures to therapeutic response and clinical evaluation of statin combination therapy
Li, Tang, Wang, Zhu, Lu, Zhang, Guo et al | August 13 2025 | Cell Stem Cell
A physiologically relevant PDAC organoid library, coupled with deep multi‑omics, revealed enrichement in protein glycosylation and cholesterol metabolisms axis driving chemoresistance and identified statins effectively targeting chemoresistant PDAC organoids. What is particularly exciting is that authors suggested a clinical benefit of a statin candidate in a phase II trial as add-on therapy, highlighting the potential of organoid library to identify possible target and treatment.
Context:
Pancreatic cancer remains stubbornly resistant to standard chemotherapy. The field needs human models that connect tumor biology to therapy response and reveal tractable resistance mechanisms.
What authors show:
Built a biobank of 260 pancreatic cancer organoid lines plus 18 normal ductal organoids from resections, EUS-FNA biopsies, and ascites; split into discovery/validation cohorts.
Ran deep multi-omics (WGS, RNA-seq, ATAC-seq, proteomics, glycoproteomics), drug and radiation sensitivity testing.
Prioritized noncoding and coding drivers and validated select hits with CRISPR in organoids.
Tested statins and downstream pathway inhibitors in organoids and xenografts; then ran a single-arm phase 2 add-on (atorvastatin 80 mg/day + ongoing chemo).
Key findings:
Functional noncoding changes and pathway analyses converged on protein glycosylation; knocking down GALNT12/18, B3GNT2, FUT1 curtailed organoid growth.
Chemoresistant organoids showed heightened glycosylation and cholesterol metabolism; statins restored chemo sensitivity, and glycan/cholesterol-pathway inhibitors synergized with FOLFIRINOX components.
In the clinical pilot, adding atorvastatin to plateauing chemo produced promising biomarker responses in ~70% of evaluable patients.
Implications:
A large, well-characterized PDAC organoid + multi-omics resource links glycosylation and cholesterol-metabolism features to chemoresistance and suggests they are drug-addressable. More broadly, it (i) highlights how noncoding regulatory regions can shape pathway expression relevant to resistance, and (ii) showcases how organoid libraries + multi-omics can surface clinically translatable candidates, exemplified by the phase II statin add-on signal.
❤️ Thank you for reading this edition of Organoids Digest. If you enjoyed it, let us know by clicking the heart button below
🌐 Visit our site to learn more about 3D cell culture and organ-model know-how: cherrybiotech.com
🔎Try our GPT Organoids, a chatbot designed to help you find the best model for your research here